Program
The MiFoBio 2025 thematic school will be held from Friday October 10 to Friday October 17 (shuttle departure).
After the 2021 post-confinement and 2023 GDR 20th anniversary editions, for which we chose to welcome more participants (390!), we’ll be welcoming a smaller number of participants (330) in 2025, while maintaining the same interdisciplinary approach to science and technology. As always, the program will be rich and varied, with a balance of lectures, seminars, round-table discussions and workshops covering a broad spectrum of topics.
What’s more, this year we’ll have the opportunity to host a European COST Action! 5 sessions will be organized as part of the week’s program as described at the bottom of this page.
Previsional schedule:
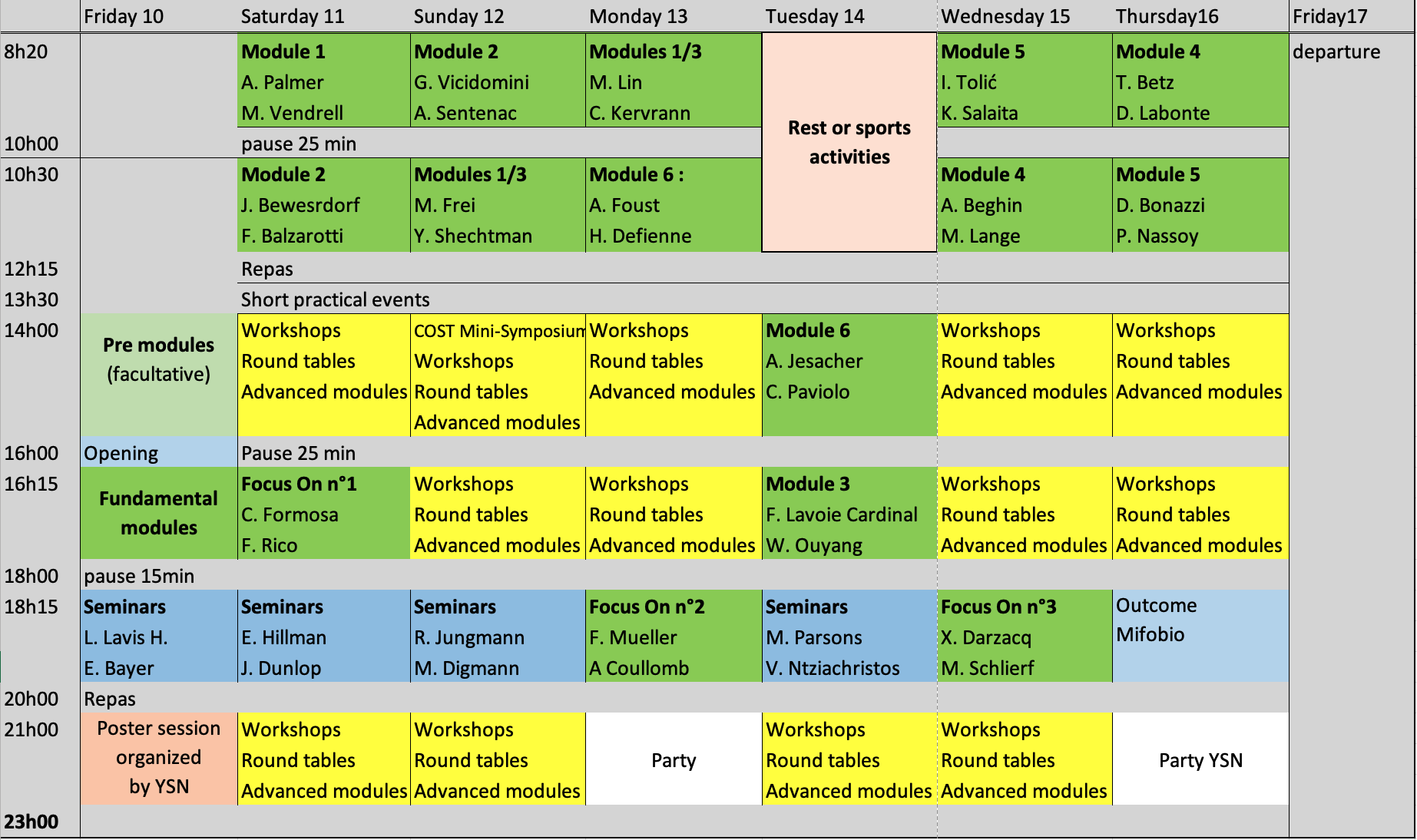
The morning coffee breaks on October 11 and 12 are sponsored by our partners Okolab and Phasics, respectively.

MiFoBio will offer the following 6 teaching modules and associated seminars:
Module 1: Marking strategies at different scales of life
Coordination : Marie Erard, Arnaud Gautier
- Michael LIN, Stanford University (USA), “New dimensions in optical imaging: Exploring space and time with fluorescent and bioluminescent proteins”
- Amy PALMER, University of Colorado Boulder (USA), “Genetically encoded sensors and how to use them quantitatively to learn new biology”
- Michelle FREI, ETH Zürich (Switzerland), “Chemigenetic kinase biosensors for multiplexed activity sensing and super-resolution imaging”
- Marc VENDRELL, University of Edinburgh (Scotland), “Fluorescent probes for translational bioimaging”
Module 2: Nanoscopy : the challenges of spatial and temporal quantification
Coordination : Sandrine Lévêque-Fort, Lydia Danglot
- Giuseppe VICIDOMINI, Istituto Italiano di Tecnologia, Genoa (Italy), “Photon-Resolved Microscopy: Image-Scanning Microscopy with SPAD Array Detector and Beyond”
- Francisco BALZAROTTI, IMP Vienna (Austria), “Introduction to MINFLUX Imaging and Tracking”
- Jörg BEWERSDORF, Yale University (USA), “Single-molecule Localization Microscopy Techniques: Foundations and Cell Biological Applications”
- Anne SENTENAC, Institut Fresnel (France), “Super-resolved fluorescence microscopy using Structured Illumination Microscopy”
Module 3: Analysis, simulation and modelling: a new paradigm with AI ?
Coordination : Aymeric Leray, Jean-Baptiste Sibarita
- Charles KERVRANN, INRIA Rennes, Institut Curie (France), “Deep learning to detect and identify molecules in 3D microscopy images“
- Flavie LAVOIE-CARDINAL, Université Laval (Quebec), “Decoding synaptic diversity with AI-assisted nanoscopy”
- Wei OUYANG, KTH Royal Institute of Technology, Stockholm (Sweden), “Generative AI for Microscopy: From Automated Imaging to Virtual Cell Modeling”
- Yoav SHECHTMAN, Technion – Israel Institute of Technology (Israel), “3D localization microscopy – spreading the point-spread function”
Module 4: Multi-scale imaging of biological systems
Coordination : Rémi Galland, Gaëlle Recher
- Merlin LANGE, Institut de la Vision, Paris, (France), “Building a Multimodal Atlas of Vertebrate Development”
- Timo BETZ, Goettingen Univ. (Germany), “Flow, Force & Form: Multimodal 3-D Microscopy for Living-Tissue Mechanics”
- David LABONTE, Imperial College London (UK), “Photogrammetry, synthetic data, and high-throughput automatic data extraction in insects”
- Anne BEGHIN, MBI, National University of Singapore (Singapore), “Breaking scale: Image Analysis–Based AI to Streamline multilevels 3D quantitative Imaging and Tackle the vEM Challenge”
Module 5: Physical measurements, control and handling – Mechanobiology
Coordination : Cécile Leduc, Renaud, Olivier
- Khalid SALAITA, Emory University, Atlanta, Georgia (USA), “May the Force Be Measured: Mechano probes reveal the molecular forces generated by cells”
- Iva TOLIĆ, Ruder Boskovic Institute, Zagreb (Croatia), “Mechanobiology of the Mitotic Spindle”
- Daria BONAZZI, Institut Pasteur / Institut Jacques Monod, Paris (France), “How Mechanical Forces Shape Bacterial Infections?”
- Pierre NASSOY, Bordeaux Univ. (France), “Formation, morphometrics and mechanics of epiblast models”
Module 6: Waves on living organisms: breakthrough strategies in life imaging
Coordination : Thomas Chaigne, Pascal Berto
- Alexander JESACHER, Medical University of Innsbruck (Austria), “Fundamentals of adaptive optics in microscopy”
- Amanda FOUST, Imperial College London (UK), “Light-field deep learning for fast, sensitive 4D imaging in scattering tissue”
- Chiara PAVIOLO, CEA-LETI, Grenoble (France), “AI-driven holography: neuronal microscopy for 2D and 3D live-cell imaging applications”
- Hugo DEFIENNE, CNRS – Sorbonne Univ., Paris (France), “Application of quantum imaging to microscopy”
Seminars:
- Maddy PARSONS, King’s College London (UK), “Imaging dynamics and mechanics in disease”
- Vasilis NTZIACHRISTOS, Technical Univ. of Munich (Germany), “Listening to light: advances in hybrid optical and optoacoustic microscopy”
- Michelle DIGMAN, Univ. of California, Irvine (USA), “Multiplexed Metabolic and Mitochondria tracking in 3D MulticellularSystems”
- Elisabeth HILLMAN, Colombia Univ. , New York (USA), “High-speed light sheet imaging – how and why ?”
- Luke LAVIS, Janelia Research Campus, HHMI – Ashburn, Virginia (USA), “Turning dyes on and off for biological imaging”
- Emmanuelle BAYER, Univ. Bordeaux (France), “How and why plant cells communicate”
- Ralf JUNGMANN, LMU and MPI of Biochemistry, Munich (Germany), “From DNA Nanotechnology to Biomedical Insight: Towards Single-Molecule Spatial Omics”
- John DUNLOP, Univ. of Salzburg (Austria), COST Action CA22153, “Geometric clues direct tissue patterning, how surface curvature influences cell behaviour”
“Focus On” :
For this edition of MiFoBio, we have implemented a new concept named “Focus On”. “Focus on” intend to illustrate applications in the field of biology of specific techniques. We will have 3 distinct sessions, the first will deal with AFM and its capabilities to deciphered molecular interactions in biological samples (FO1). The second will show how using multiplexed image of the transcriptome will can gain information about the biological state, from the cell to the tissue (FO2). Finally, recent applications of single molecule fluorescence microscopy in the field of drug screening will be discussed (F03).
FO1 : “Molecular interaction and Mechanosensing using Atomic Force Microscopy”
Cécile FORMOSA-DAGUE, Toulouse (France) ; Felix RICO, Marseille (France)
FO2 : “Multiplexed Imaging of the Transcriptome — From Single Molecules to Tissues”
Florian MUELLER, Institut Pasteur, Paris (France) ; Alexis COULLOMB, Toulouse (France)
FO3 : “Single Molecule Fluorescence Microscopies for Drug Discovery”
Xavier DARZACQ, Berkeley (USA) ; Michael SCHLIERF, Dresden (Germany)
“Premodules for beginners” :
These courses are designed for people who do not have a basic knowledge of the relevant fields. These are optional courses, for which there will not be a huge number of places (maximum 15 people per course). Mifobio participants wishing to register will be invited to do so when they receive, directly by e-mail, a form designed to collect a set of information for the organization (arrival and departure times, accommodation, and therefore possible registration for a pre-module). These courses will a priori be given in French. The description is available below:
Cell biology basics – Delphine Muriaux
The organism, whether plant or animal, is made up of billions of cells of different types and functions, which differentiate during development following fertilization. In this course, we will pay particular attention to the cell as a unit, defining its molecular aspects in general terms. We’ll describe the differences between eukaryotic and prokaryotic cells, their intracellular compartments (Nucleus, Reticulum, Golgi, Mitochondria, Endosomes…) and the notions of Membranes, Proteins, RNA and DNA, in size, numbers and color. Finally, we’ll look at transfection, transduction and infection, as tools for expressing fluorescent proteins in these cells and visualizing them by microscopy.
The organization of living organisms is constrained by the laws of chemistry and physics, and its study requires tools whose dynamic ranges and limits must be adjusted to the right dimensions. The second part of this course introduces the notions of space and time of processes from the molecular to the organismal scale.
Basic optics – Olivier Haeberlé
This pre-module introduces the basics of image formation in a conventional optical microscope and places them in the context of the various technologies that will be covered in greater depth in the modules (elementary geometrical optics, phase and polarization, diffractive optics and PSF, introductions to super-resolution and microtomography techniques).
Basic photophysics and chemistry for microscopy – Dominique Bourgeois
This pre-module provides an overview of the various processes involved in light absorption and fluorescence, focusing on the properties of fluorophores useful in microscopy: absorption spectra, molar absorption coefficient, emission spectra, quantum yield, brightness, lifetime, fluorescence versus phosphorescence, reactions with the environment, bleaching, nonlinear optics and two-photon absorption, second harmonic generation). We’ll also take a brief look at the chemical reactions induced by light absorption (photoisomerization and decaging). The thread running through this pre-module will be a fluorescent probe that will be synthesized at MIFOBIO in the Chem-Lab, and we’ll be looking at the various stages involved in its design, synthesis, chemical and photophysical characterization, so that it can be used as a mitochondrial probe on the systems present at MIFOBIO.
Fundamentals of image analysis: from classical methods to machine learning – Aymeric Leray
In this pre-module, we will begin by reviewing the concept of digital images (dynamics, noise, format) before moving on to present the basic techniques of image preprocessing and segmentation (thresholding, filters, binarization, and morphological analysis). We will conclude by introducing more recent approaches based on artificial intelligence and machine learning (decision trees, convolutional neural networks). We will explain the principles behind these methods and illustrate them with concrete examples to provide an initial overview of their possibilities.
FLIM, FRET, FCS: F-microscopies for the dynamics and interactions of macromolecules in living cells – Marc Tramier
Quantitative fluorescence microscopy methods such as FLIM (Fluorescence Lifetime Imaging Microscopy), FRET (Forster Resonance Energy Transfer), and FCS (Fluorescence Correlation Spectroscopy), also known as F-microscopies, are becoming increasingly accessible with the development of commercial instruments that are ever easier to use. They enable the measurement of proximity and diffusion (or co-diffusion) of macromolecules labeled with fluorophores in living cells, making them techniques of choice in cell biology. In this course, we will discuss these methods and show how it is becoming increasingly easy to use them in microscopy to study the spatiotemporal regulation of protein activity and interactions.
COST action – CA22153 – European Curvature and Biology Network (EuroCurvoBioNet)
The “EuroCurvoBioNet” COST Action brings together biologists, physicists, mathematicians, designers and others to elucidate the role of curvature in biological systems. The integration of this action into Mifobio should open up discussion about problem solving, existing tools and fundamental questions related to the interaction between curvature and biology, as well as the imaging of curved surfaces.
5 COST Action sessions have been organized as part of the MiFoBio program, according to the schedule below:
- Saturday, October 11th at 6pm Seminar: John Dunlop, Paris-Lodron-University of Salzburg, Austria: “Geometric clues direct tissue patterning, how surface curvature influences cell behaviour”.
- Sunday, October 12th at 2pm Mini-symposium “EuroCurvoBioNet COST Action: the role of curvature on Biology, Tissue Engineering, Regenerative Medicine, and Sustainable Materials”: we will introduce the COST (CA22153) objectives through the presentations of representative research projects of the working groups of the Action:
2-2.15 pm Introduction
2.15-2.45 pm Clémentine Ferrari, Max Planck Institute of Colloids and Interfaces, Germany:“Mechanobiology of Biofilms: Influence of surface micro-topography on biofilm macro-morphology”
2.45-3.15 pm André Castro, IDMEC, Instituto Superior Técnico and ESTSetúbal, Instituto Politécnico de Setúbal, Portugal: “On the design and modelling of TPMS scaffolds for tissue engineering”
3.15-3.45 pm Łucja Kowalewska, University of Warsaw, Poland: “Complex Nanoscale Topologies in Plants – Diamond and Gyroid Membrane Networks in Plastids”
- Sunday, October 12th at 9pm: Round table “EuroCurvoBioNet COST Action: exploring new approaches to investigate and characterize cellular responses to curved environments at different scales”: we will discuss about the key fundamental questions to address in order to better understand the role of curvature on cell behavior and we will propose an interactive discussion about the needs and issues encountered in producing, imaging and analyzing curved surfaces in vitro.
- Monday, October 13th at 2pm and Tuesday, October 14th at 9pm: Workshop”Development of microstructured cell culture substrates from 3D models using grayscale lithography”
- Wednesday-Thursday, October 15-16th at 2pm: Workshop “Epithelium monolayers deformed by curved substrates: advantages and drawbacks of light-sheet and confocal microscopy”
